


I. Prepare the hardware for the experiment:
-
Turn on the computer and all the devices: Cellsorter Piezo control unit, (microscope, camera, control units for motorized components).
-
Assemble the Piezo Head and fill it with filtered milli-Q water. Further details of this process are shown in the video tutorial for the Piezo Head.
-
Mount the CellSorter sample holder aluminum insert into the motorized stage.
-
Insert the Petri dish with your sample (cells covered by 3 mL medium) and fix the dish with the magnetic cap. Or use a Petri dish marked with permanent marker and 3 mL water in it to set up microscope focus later.

Figure 1. Prepare the hardware
II. Setup window
-
Launch the Cellsorter software.
-
As shown below, we have the opportunity to work in 4 menu items: SETUP, MANUAL, PROTOCOL, EXECUTE
-
On the first window (Fig. 2.) it is possible to check the settings of the stage, focus, manipulator, camera, illumination, fluidics, and the sample holder.
-
At the sample holder tab select the sample holder placed in the stage, as well as the size of the piezo head, with the corresponding O-ring, at the fluidics tab.
-
More detailed settings are available in the MANUAL window.

Figure 2. Setup window.
III. Manual window
Calibration of the stage and camera
-
Calibrate the injection positions by bringing the calibration crosshair into the middle of the camera field of view. We have two options for this calibration in the stage tab. “Manual calib” or “Autocalib”. (Fig. 3.1.)

Figure 3.1. Settings of the stage
2. Set up camera properties in the Camera control window (Exposition time, resolution, binning, etc.)
3. Set and save Presets. These presets include Camera and Light setups. (Fig. 3.2.)

Figure 3.2. Setting and saving Presets
4. Also, here you can determine what color the marking should be if a fluorescent light source is used. (Fig. 3.3.)

Figure 3.3. Application of fluorescent light

Figure 3.4. Image of beads excited by a fluorescent light source.
Two option is available for the calibration of the micropipette: manual or automatic
A. Manual calibration of micropipette:
5. Move down the micropipette with the motorized micromanipulator using the joystick or the Micromanipulator window in the software. Reach the surface of the water in the Petri dish with the tip. Now the tip is ~3 mm above the bottom of the dish. (Fig. 3.5.)

Figure 3.5. Calibration of the manipulator
6. Position the micropipette exactly above the center of the objective lens using the manual screws of the manipulator. (Fig. 3.6.)


Figure 3.6. Manual screws of the manipulator for NanoPick and NanoPick1000 system
7. Lift the objective by 1 mm to prevent a sudden collision of the needle with the surface of the petri dish.
8. Move down the micropipette gradually in steps starting with 1000 um, then 500 um, then 100 um. When the micropipette tip is almost sharp use 10 um steps.
9. Move the lens down by 1mm, back to the sharp image,
10. Move the needle down also by about 1mm.
11. Find where the image of the needle is sharp.
12. The goal is to find the point where the pipette contacts the surface of the petri dish. (here the image will be sharp, then we can provide the necessary data to pick up the cells during the experiment) (Fig. 3.7.)
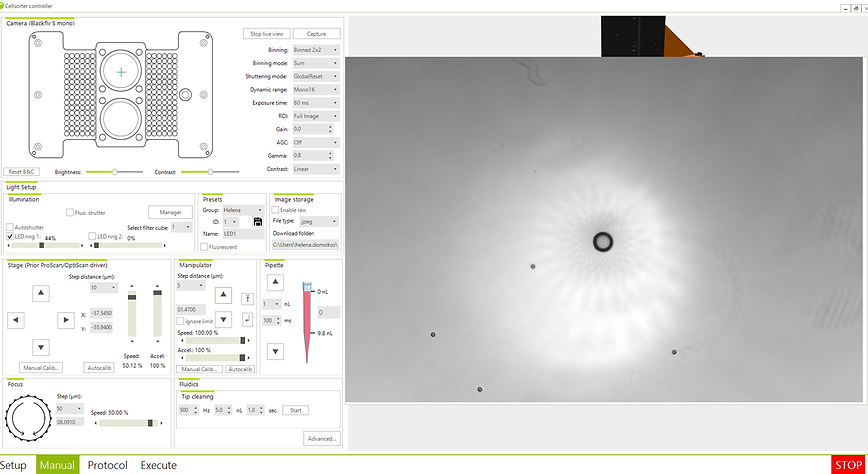
Figure 3.7. Pipette calibration, when the image of the beads and the tip of the pipette is sharp
13. This can be done by gently moving the manipulator to the side, or with the semi-automatic mover, which moves the manipulator down by 5 um and to the side by 10 um. (Fig. 3.8.)

Figure 3.8. The semi-automatic mover, of the manipulator
14. Finalizing the pipette calibration: by clicking the “Manual Calib” button a window will appear whereby selecting the box “Set manipulator position to 0” will reset the Z coordinate of the manipulator, and before that, it is important to move the crosshair to the center of the pipette tip shown in the picture. (Fig. 3.9.)

Figure 3.9. Set manipulator Z position to 0 after tip calibration
B. Automatic calibration of the micropipette:
1.In this case, we need a length calibrated micropipette. Length of the micropipette is written on the box.
2. Select the "Autocalib" option for the manipulator, and follow the instructions:
-
First of all, you need to specify the parameters of the pipette (length, tip diameter) (Figure 3.10a)
-
Make sure that the sample (or the reference dish used for calibration) is properly set for imaging as detailed in Figure 3.10b.


Figure 3.10. a, Parameters of the pipette b, Requirements for automatic pipette calibration
3. The device then automatically performs the steps detailed above in manual calibration. Major steps are listed below:

Figure 3.11. The pipette goes to the upper home position and starts the calibration from there

Figure 3.12. Starts detecting the pipette tip by lifting the objective
4. If the "touch the surface" option was selected, it finds the contact height and finally sets the pipette at a height of 5 um above the surface

Figure 3.13. Finding the contact height
4. At the end of the calibration, it sets the position (Z coordinate) of the manipulator to 0, if this was also requested at the beginning of the process.

Figure 3.14. Pipette calibration completed
IV. Protocol window
1. Focus the microscope on the cells and find your region of interest.

Figure 4.1. PROTOCOL window, which include SCANNING, ANALYZING and SORTING steps.
Scanning
2. Use the Scanning tab to scan your region of interest. Set up the top left and the bottom right corner of the region. (Fig. 4.2.)

Figure 4.2. Scan area settings
Notice: If a scan has already been performed and you click on the sign, it will also change the z coordinate (the focus).
3. Give your image resolution in the Scanning window. You can also choose from a drop-down list. Please note that camera binning and objective magnification (which was previously set in manual windows) will affect the image resolution. (Fig. 4.2.)
4. It is important to make sure that the pipette is at a safe height during operations, for example when moving from one Petri dish to another, or during scanning. The “Lift pipette before scanning” feature helps in this.
(Fig. 4.3.)

Figure 4.3. Scanning setups
5. Compensate if the Petri dish is tilted by pushing the ‘Set sharpest plane’ button. (Fig. 4.3.)
6. Follow the directions given there. (Use the live view image from the camera window to focus the microscope onto the corners of the region of interest.) (Fig. 4.4.)

Figure 4.4. Figure Settings required for uneven surfaces. The manipulator will use the data saved here to correct the picking up of the cells even on an uneven surface.
7. In the Channels tab you can select previously saved presets from the drop-down menu. This determines in which channel (camera and light settings) the scanning will take place.
8. Image background correction can be performed.

Figure 4.5. Here you can select how many and in which channels the scanning should take place.
Analyzing
9. Analyze the scanned area in the Analyzing window. Detect the cells automatically by software or manually using the mouse. You can tune the cell recognition of the software by adjusting the detection parameters. (Sensitivity, brightness, cell area). Always start from lower sensitivity and increase detection sensitivity gradually.
10. Push the ‘Detect cells in all images’ button. This will recognize cells in all images of the scanned region.
11. You can also remove or add cells manually after an automated detection.

Figure 4.6. Detection of the cells in the scanning area.
Sorting
General tab:
12. You can visualize the path of the micropipette and order of cells to be picked up. (Fig.4.7.)
13. The preset can be selected separately for the each steps of the protocol. Click on the desired step in the list, then select the preset from the "Preset for selected image type" drop-down menu.

Figure 4.7. Sorting: General tab
Notice: If you are not using the 10x objective (but a higher one), the following setting may be necessary:
Sorting – general:
-
move objective down after pick-up
-
move objective up before depo: (as well as setting the z (stage) parameter of the reservoir to a lower level)
Pick Up tab:
14. It is important to set the distance of the micropipette’s tip and the dish for picking up the cells from the surface: “Low position” setting. (Fig.4.8.)
15. "Adaptive cell targeting" corrects the coordinates of cells if they have moved after scanning:

Figure 4.8. Sorting: Pick up tab.
Deposition:
16. You can choose to deposit the cells into PCR tubes or into the second petri dish, or to a manually selected area. (Fig.4.9.)
17. The location of the deposition is indicated by a yellow rectangle. (Fig. 4.10.)

Figure 4.9. Sorting: Deposition tab

Figure 4.10. Sorting: Deposition area in the second petri dish
Piezo:
18. The deposition height is the distance between the micropipette’s tip and the dish when depositing cells. In the case of the reservoir, the manipulator must be set to the appropriate position and then saved in the Piezo tab "read reservoir stage, and manipulator position". With the arrow highlighted below, the manipulator moves the micropipette to a height of 1 mm. The up and down arrows next to it move the manipulator by 10 µm. (Fig.4.11.)

Figure 4.11. Reservoir coordinates
19. Operation of the piezoelectric micropipette during the sorting process can be set here. Pick-up, deposition and reset (in the Reservoir) fluid volumes, and timings. (Fig.4.12.)

Figure 4.12. Sorting: Piezo tab
V. Execute window
1. Start sorting from the EXECUTE window.





